knitr::opts_chunk$set(echo = TRUE)
library(tidyverse)
library(broom)
library(prettydoc)
library(corrplot)
library(ggformula)
library(palmerpenguins) # Allison Horst's `penguins` data.
##
library(tidymodels)
library(dials)
library(modeldata)
library(rsample)
library(recipes)
library(yardstick)
library(parsnip)Random Forests
Penguin Random Forest Model withrandomForest
Using the penguins dataset and Random Forest Classification.
penguinsspecies <fct> | island <fct> | bill_length_mm <dbl> | bill_depth_mm <dbl> | flipper_length_mm <int> | body_mass_g <int> | sex <fct> | year <int> |
|---|---|---|---|---|---|---|---|
| Adelie | Torgersen | 39.1 | 18.7 | 181 | 3750 | male | 2007 |
| Adelie | Torgersen | 39.5 | 17.4 | 186 | 3800 | female | 2007 |
| Adelie | Torgersen | 40.3 | 18.0 | 195 | 3250 | female | 2007 |
| Adelie | Torgersen | NA | NA | NA | NA | NA | 2007 |
| Adelie | Torgersen | 36.7 | 19.3 | 193 | 3450 | female | 2007 |
| Adelie | Torgersen | 39.3 | 20.6 | 190 | 3650 | male | 2007 |
| Adelie | Torgersen | 38.9 | 17.8 | 181 | 3625 | female | 2007 |
| Adelie | Torgersen | 39.2 | 19.6 | 195 | 4675 | male | 2007 |
| Adelie | Torgersen | 34.1 | 18.1 | 193 | 3475 | NA | 2007 |
| Adelie | Torgersen | 42.0 | 20.2 | 190 | 4250 | NA | 2007 |
summary(penguins) species island bill_length_mm bill_depth_mm
Adelie :152 Biscoe :168 Min. :32.10 Min. :13.10
Chinstrap: 68 Dream :124 1st Qu.:39.23 1st Qu.:15.60
Gentoo :124 Torgersen: 52 Median :44.45 Median :17.30
Mean :43.92 Mean :17.15
3rd Qu.:48.50 3rd Qu.:18.70
Max. :59.60 Max. :21.50
NA's :2 NA's :2
flipper_length_mm body_mass_g sex year
Min. :172.0 Min. :2700 female:165 Min. :2007
1st Qu.:190.0 1st Qu.:3550 male :168 1st Qu.:2007
Median :197.0 Median :4050 NA's : 11 Median :2008
Mean :200.9 Mean :4202 Mean :2008
3rd Qu.:213.0 3rd Qu.:4750 3rd Qu.:2009
Max. :231.0 Max. :6300 Max. :2009
NA's :2 NA's :2 | Name | Piped data |
| Number of rows | 344 |
| Number of columns | 8 |
| _______________________ | |
| Column type frequency: | |
| factor | 3 |
| numeric | 5 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| species | 0 | 1.00 | FALSE | 3 | Ade: 152, Gen: 124, Chi: 68 |
| island | 0 | 1.00 | FALSE | 3 | Bis: 168, Dre: 124, Tor: 52 |
| sex | 11 | 0.97 | FALSE | 2 | mal: 168, fem: 165 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| bill_length_mm | 2 | 0.99 | 43.92 | 5.46 | 32.1 | 39.23 | 44.45 | 48.5 | 59.6 | ▃▇▇▆▁ |
| bill_depth_mm | 2 | 0.99 | 17.15 | 1.97 | 13.1 | 15.60 | 17.30 | 18.7 | 21.5 | ▅▅▇▇▂ |
| flipper_length_mm | 2 | 0.99 | 200.92 | 14.06 | 172.0 | 190.00 | 197.00 | 213.0 | 231.0 | ▂▇▃▅▂ |
| body_mass_g | 2 | 0.99 | 4201.75 | 801.95 | 2700.0 | 3550.00 | 4050.00 | 4750.0 | 6300.0 | ▃▇▆▃▂ |
| year | 0 | 1.00 | 2008.03 | 0.82 | 2007.0 | 2007.00 | 2008.00 | 2009.0 | 2009.0 | ▇▁▇▁▇ |
# library(corrplot)
cor <- penguins %>%
select(where(is.numeric)) %>%
cor()
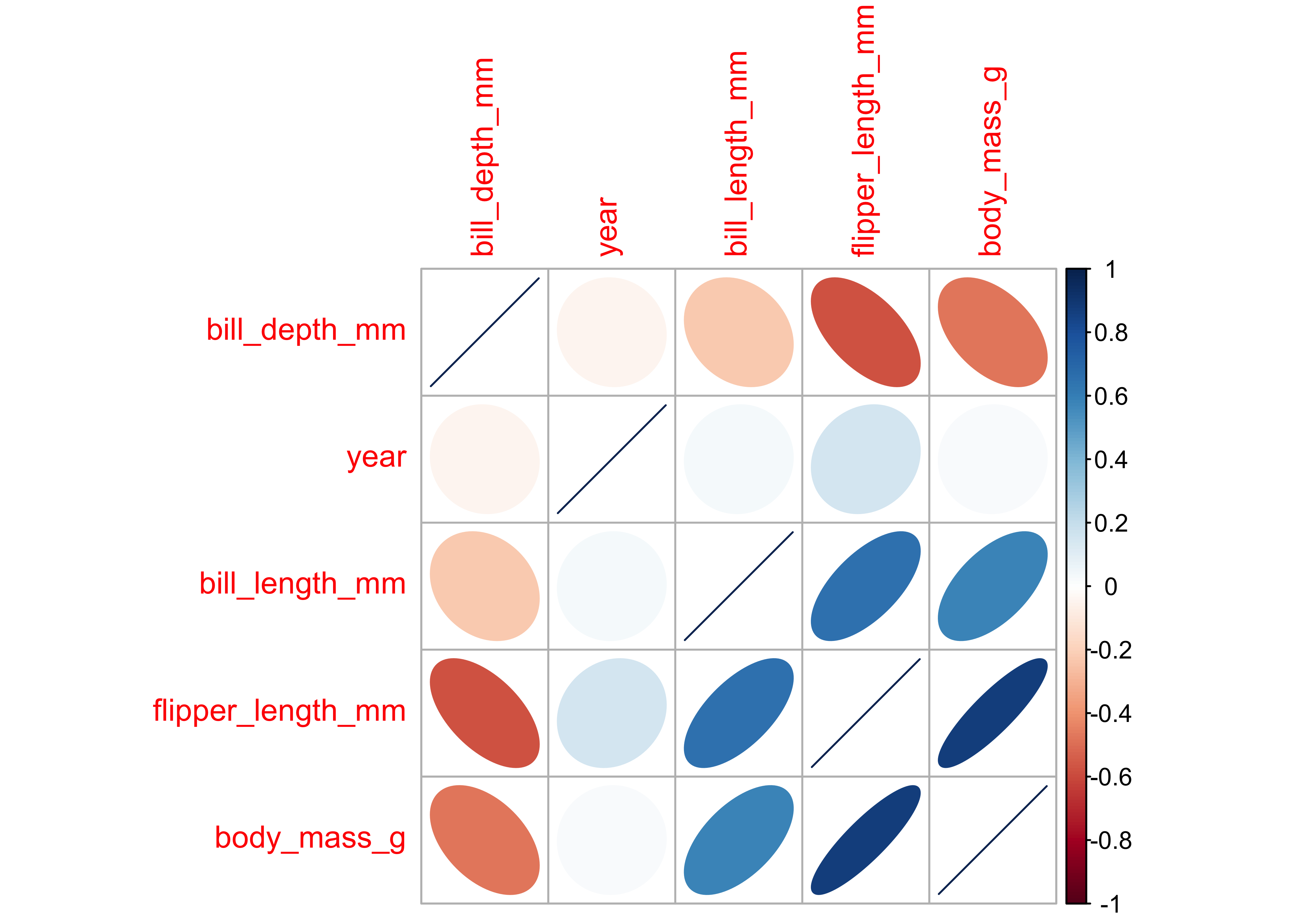
cor %>% corrplot(., method = "ellipse", order = "hclust", tl.cex = 1.0, )# try these too:
# cor %>% corrplot(., method = "square", order = "hclust",tl.cex = 0.5)
# cor %>% corrplot(., method = "color", order = "hclust",tl.cex = 0.5)
# cor %>% corrplot(., method = "shade", order = "hclust",tl.cex = 0.5)Notes: - flipper_length_mm and culmen_depth_mm are negatively correlated at approx (-0.7) - flipper_length_mm and body_mass_g are positively correlated at approx 0.8
So we will use steps in the recipe to remove correlated variables.
Penguin Data Sampling and Recipe
# Data Split
penguin_split <- initial_split(penguins, prop = 0.6)
penguin_train <- training(penguin_split)
penguin_test <- testing(penguin_split)
penguin_split<Training/Testing/Total>
<199/134/333>head(penguin_train)species <fct> | island <fct> | bill_length_mm <dbl> | bill_depth_mm <dbl> | flipper_length_mm <int> | body_mass_g <int> | sex <fct> | year <int> |
|---|---|---|---|---|---|---|---|
| Gentoo | Biscoe | 47.5 | 14.0 | 212 | 4875 | female | 2009 |
| Gentoo | Biscoe | 46.2 | 14.5 | 209 | 4800 | female | 2007 |
| Adelie | Dream | 36.0 | 17.8 | 195 | 3450 | female | 2009 |
| Chinstrap | Dream | 46.4 | 18.6 | 190 | 3450 | female | 2007 |
| Gentoo | Biscoe | 42.6 | 13.7 | 213 | 4950 | female | 2008 |
| Gentoo | Biscoe | 45.1 | 14.5 | 215 | 5000 | female | 2007 |
# Recipe
penguin_recipe <- penguins %>%
recipe(species ~ .) %>%
step_normalize(all_numeric()) %>% # Scaling and Centering
step_corr(all_numeric()) %>% # Handling correlated variables
prep()
# Baking the data
penguin_train_baked <- penguin_train %>%
bake(object = penguin_recipe, new_data = .)
penguin_test_baked <- penguin_test %>%
bake(object = penguin_recipe, new_data = .)
head(penguin_train_baked)island <fct> | bill_length_mm <dbl> | bill_depth_mm <dbl> | flipper_length_mm <dbl> | body_mass_g <dbl> | sex <fct> | year <dbl> | species <fct> |
|---|---|---|---|---|---|---|---|
| Biscoe | 0.6413275 | -1.6071541 | 0.7871873 | 0.8295204 | female | 1.1783814 | Gentoo |
| Biscoe | 0.4036096 | -1.3532485 | 0.5731427 | 0.7363777 | female | -1.2818130 | Gentoo |
| Dream | -1.4615611 | 0.3225288 | -0.4257325 | -0.9401915 | female | 1.1783814 | Adelie |
| Dream | 0.4401816 | 0.7287778 | -0.7824736 | -0.9401915 | female | -1.2818130 | Chinstrap |
| Biscoe | -0.2546859 | -1.7594975 | 0.8585356 | 0.9226631 | female | -0.0517158 | Gentoo |
| Biscoe | 0.2024638 | -1.3532485 | 1.0012320 | 0.9847583 | female | -1.2818130 | Gentoo |
Penguin Random Forest Model
penguin_model <-
rand_forest(trees = 100) %>%
set_engine("randomForest") %>%
set_mode("classification")
penguin_modelRandom Forest Model Specification (classification)
Main Arguments:
trees = 100
Computational engine: randomForest parsnip model object
Call:
randomForest(x = maybe_data_frame(x), y = y, ntree = ~100)
Type of random forest: classification
Number of trees: 100
No. of variables tried at each split: 2
OOB estimate of error rate: 2.01%
Confusion matrix:
Adelie Chinstrap Gentoo class.error
Adelie 86 2 0 0.02272727
Chinstrap 2 38 0 0.05000000
Gentoo 0 0 71 0.00000000# iris_ranger <-
# rand_forest(trees = 100) %>%
# set_mode("classification") %>%
# set_engine("ranger") %>%
# fit(Species ~ ., data = iris_training_baked)Metrics for the Penguin Random Forest Model
# Predictions
predict(object = penguin_fit, new_data = penguin_test_baked) %>%
dplyr::bind_cols(penguin_test_baked) %>%
glimpse()Rows: 134
Columns: 9
$ .pred_class <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…
$ island <fct> Torgersen, Torgersen, Torgersen, Biscoe, Biscoe, Bis…
$ bill_length_mm <dbl> -1.3335592, -1.7541369, 0.3670377, -1.0592694, -0.62…
$ bill_depth_mm <dbl> 1.08424573, 0.62721557, 2.20143056, 0.47487218, 0.72…
$ flipper_length_mm <dbl> -0.56842897, -1.21056301, -0.49708074, -1.13921478, …
$ body_mass_g <dbl> -0.940191505, -1.095429393, -0.008764181, -0.3192399…
$ sex <fct> female, female, male, male, male, male, female, fema…
$ year <dbl> -1.2818130, -1.2818130, -1.2818130, -1.2818130, -1.2…
$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…# Prediction Accuracy Metrics
predict(object = penguin_fit, new_data = penguin_test_baked) %>%
dplyr::bind_cols(penguin_test_baked) %>%
yardstick::metrics(truth = species, estimate = .pred_class).metric <chr> | .estimator <chr> | .estimate <dbl> | ||
|---|---|---|---|---|
| accuracy | multiclass | 0.9701493 | ||
| kap | multiclass | 0.9531632 |
# Prediction Probabilities
penguin_fit_probs <-
predict(penguin_fit, penguin_test_baked, type = "prob") %>%
dplyr::bind_cols(penguin_test_baked)
glimpse(penguin_fit_probs)Rows: 134
Columns: 11
$ .pred_Adelie <dbl> 0.99, 0.99, 0.59, 1.00, 1.00, 1.00, 0.84, 0.95, 0.92…
$ .pred_Chinstrap <dbl> 0.01, 0.01, 0.39, 0.00, 0.00, 0.00, 0.16, 0.05, 0.08…
$ .pred_Gentoo <dbl> 0.00, 0.00, 0.02, 0.00, 0.00, 0.00, 0.00, 0.00, 0.00…
$ island <fct> Torgersen, Torgersen, Torgersen, Biscoe, Biscoe, Bis…
$ bill_length_mm <dbl> -1.3335592, -1.7541369, 0.3670377, -1.0592694, -0.62…
$ bill_depth_mm <dbl> 1.08424573, 0.62721557, 2.20143056, 0.47487218, 0.72…
$ flipper_length_mm <dbl> -0.56842897, -1.21056301, -0.49708074, -1.13921478, …
$ body_mass_g <dbl> -0.940191505, -1.095429393, -0.008764181, -0.3192399…
$ sex <fct> female, female, male, male, male, male, female, fema…
$ year <dbl> -1.2818130, -1.2818130, -1.2818130, -1.2818130, -1.2…
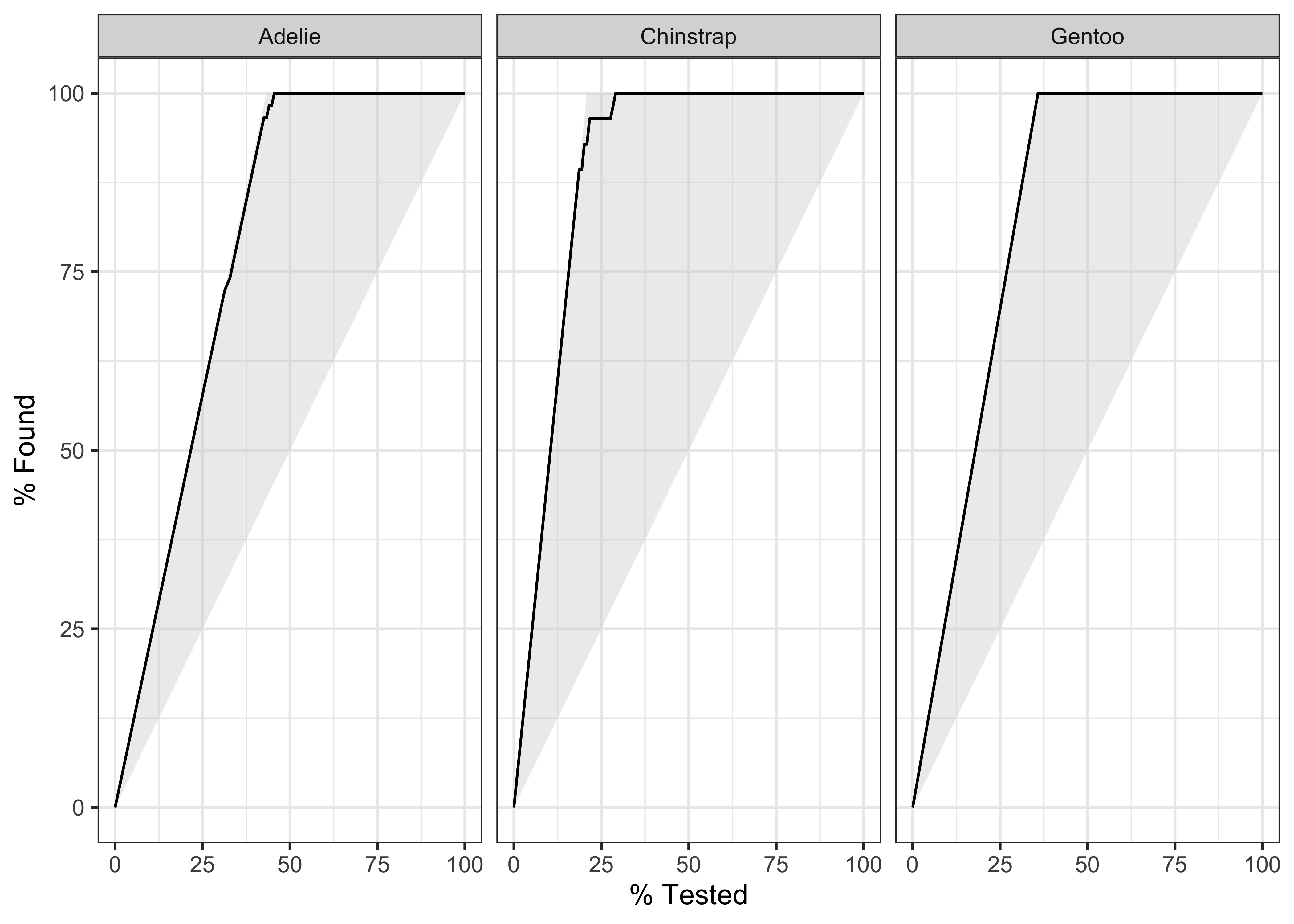
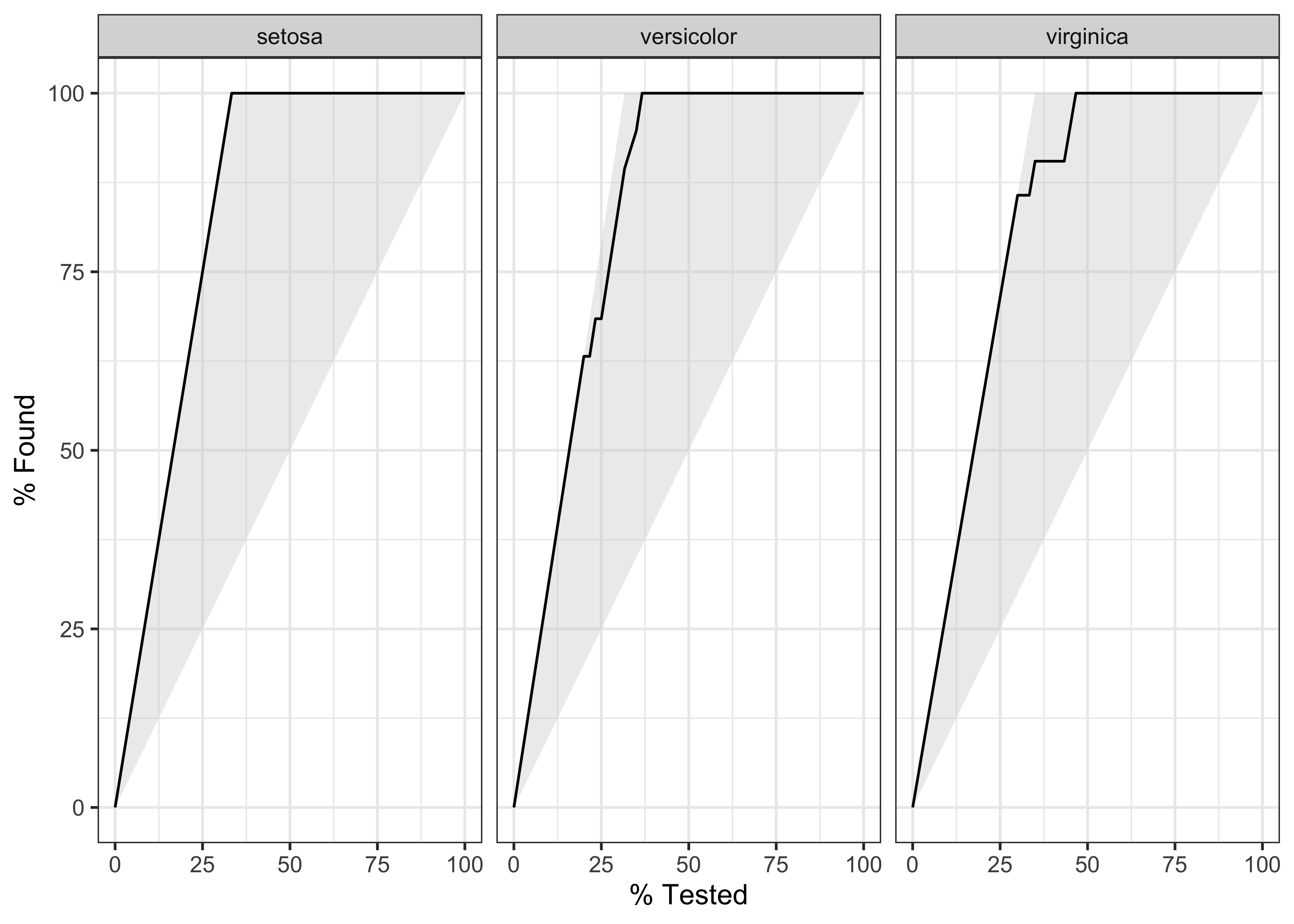
$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…# Gain Curves
penguin_fit_probs %>%
yardstick::gain_curve(species, .pred_Adelie:.pred_Gentoo) %>%
autoplot()Using broom on the penguin model
penguin_split<Training/Testing/Total>
<199/134/333># Following do not work for `random forest models` !! ;-()
# penguin_model %>% tidy()
# penguin_fit %>% tidy()
penguin_model %>% str()List of 7
$ args :List of 3
..$ mtry : language ~NULL
.. ..- attr(*, ".Environment")=<environment: R_EmptyEnv>
..$ trees: language ~100
.. ..- attr(*, ".Environment")=<environment: R_EmptyEnv>
..$ min_n: language ~NULL
.. ..- attr(*, ".Environment")=<environment: R_EmptyEnv>
$ eng_args : Named list()
..- attr(*, "class")= chr [1:2] "quosures" "list"
$ mode : chr "classification"
$ user_specified_mode : logi TRUE
$ method : NULL
$ engine : chr "randomForest"
$ user_specified_engine: logi TRUE
- attr(*, "class")= chr [1:2] "rand_forest" "model_spec"penguin_test_bakedIris Random Forest Model with ranger
Using the iris dataset and Random Forest Classification. This part uses rsample to split the data and the recipes to prep the data for model making.
# set.seed(100)
iris_split <- rsample::initial_split(iris, prop = 0.6)
iris_split<Training/Testing/Total>
<90/60/150>Rows: 90
Columns: 5
$ Sepal.Length <dbl> 7.2, 5.1, 6.7, 6.7, 7.1, 5.0, 5.1, 5.2, 6.0, 5.9, 6.6, 5.…
$ Sepal.Width <dbl> 3.6, 3.8, 3.0, 3.1, 3.0, 3.5, 3.5, 2.7, 2.2, 3.0, 2.9, 3.…
$ Petal.Length <dbl> 6.1, 1.5, 5.2, 4.4, 5.9, 1.6, 1.4, 3.9, 5.0, 4.2, 4.6, 1.…
$ Petal.Width <dbl> 2.5, 0.3, 2.3, 1.4, 2.1, 0.6, 0.2, 1.4, 1.5, 1.5, 1.3, 0.…
$ Species <fct> virginica, setosa, virginica, versicolor, virginica, seto…Rows: 60
Columns: 5
$ Sepal.Length <dbl> 4.6, 5.4, 4.4, 4.8, 4.8, 5.7, 4.6, 4.8, 5.0, 5.2, 4.8, 5.…
$ Sepal.Width <dbl> 3.1, 3.9, 2.9, 3.4, 3.0, 3.8, 3.6, 3.4, 3.4, 3.4, 3.1, 3.…
$ Petal.Length <dbl> 1.5, 1.7, 1.4, 1.6, 1.4, 1.7, 1.0, 1.9, 1.6, 1.4, 1.6, 1.…
$ Petal.Width <dbl> 0.2, 0.4, 0.2, 0.2, 0.1, 0.3, 0.2, 0.2, 0.4, 0.2, 0.2, 0.…
$ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, setosa, s…Iris Data Pre-Processing: Creating the Recipe
The recipes package provides an interface that specializes in data pre-processing. Within the package, the functions that start, or execute, the data transformations are named after cooking actions. That makes the interface more user-friendly. For example:
recipe()- Starts a new set of transformations to be applied, similar to theggplot()command. Its main argument is the model’sformula.prep()- Executes the transformations on top of the data that is supplied (typically, the training data). Each data transformation is astep()function. ( Recall what we did with thecaretpackage: Centering, Scaling, Removing Correlated variables…)
Note that in order to avoid data leakage (e.g: transferring information from the train set into the test set), data should be “prepped” using the train_tbl only. https://towardsdatascience.com/modelling-with-tidymodels-and-parsnip-bae2c01c131c CRAN: The idea is that the preprocessing operations will all be created using the training set and then these steps will be applied to both the training and test set.
# Pre Processing the Training Data
iris_recipe <-
training(iris_split) %>% # Note: Using TRAINING data !!
recipe(Species ~ .) # Note: Outcomes ~ Predictors !!
# The data contained in the `data` argument need not be the training set; this data is only used to catalog the names of the variables and their types (e.g. numeric, etc.).Q: How does the recipe “figure” out which are the outcomes and which are the predictors? A.The recipe command defines Outcomes and Predictors using the formula interface. Not clear how this recipe “figures” out which are the outcomes and which are the predictors, when we have not yet specified them…
Q. Why is the recipe not agnostic to data set? Is that a meaningful question? A. The use of the training set in the recipe command is just to declare the variables and specify the roles of the data, nothing else. Roles are open-ended and extensible. From https://cran.r-project.org/web/packages/recipes/vignettes/Simple_Example.html :
This document demonstrates some basic uses of recipes. First, some definitions are required: - variables are the original (raw) data columns in a data frame or tibble. For example, in a traditional formula Y ~ A + B + A:B, the variables are A, B, and Y. - roles define how variables will be used in the model. Examples are:
predictor(independent variables),response, andcase weight. This is meant to be open-ended and extensible. - terms are columns in a design matrix such as A, B, and A:B. These can be other derived entities that are grouped, such as a set ofprincipal componentsor a set of columns, that define abasis functionfor a variable. These are synonymous withfeaturesin machine learning. Variables that havepredictorroles would automatically be maineffect terms.
# Apply the transformation steps
iris_recipe <- iris_recipe %>%
step_corr(all_predictors()) %>%
step_center(all_predictors(), -all_outcomes()) %>%
step_scale(all_predictors(), -all_outcomes()) %>%
prep()This has created the recipe() and prepped it too. We now need to apply it to our datasets:
- Take
trainingdata andbake()it to prepare it for modelling. - Do the same for the
testingset.
Iris Model Training using parsnip
Different ML packages provide different interfaces (APIs ) to do the same thing (e.g random forests). The tidymodels package provides a consistent interface to invoke a wide variety of packages supporting a wide variety of models.
The parsnip package is a successor to caret.
To model with parsnip: 1. Pick a model : 2. Set the engine 3. Set the mode (if needed): Classification or Regression
Check here for models available in parsnip.
Mode: classification and regression in
parsnip, each using a variety of models. ( Which Way). This defines the form of the output.Engine: The
engineis the R package that is invoked byparsnipto execute the model. E.gglm,glmnet,keras.( How )parsnipprovides wrappers for models from these packages.Model: is the specific technique used for the modelling task. E.g
linear_reg(),logistic_reg(),mars,decision_tree,nearest_neighbour…(What model).
and models have: - hyperparameters: that are numerical or factor variables that tune the model ( Like the alpha beta parameters for Bayesian priors)
We can use the random forest model to classify the iris into species. Here Species is the Outcome variable and the rest are predictor variables. The random forest model is provided by the ranger package, to which tidymodels/parsnip provides a simple and consistent interface.
library(ranger)
iris_ranger <-
rand_forest(trees = 100) %>%
set_mode("classification") %>%
set_engine("ranger") %>%
fit(Species ~ ., data = iris_training_baked)ranger can generate random forest models for classification, regression, survival( time series, time to event stuff). Extreme Forests are also supported, wherein all points in the dataset are used ( instead of bootstrap samples) along with feature bagging. We can also run the same model using the randomForest package:
library(randomForest, quietly = TRUE)
iris_rf <-
rand_forest(trees = 100) %>%
set_mode("classification") %>%
set_engine("randomForest") %>%
fit(Species ~ ., data = iris_training_baked)Iris Predictions
The predict() function run against a parsnip model returns a prediction tibble. By default, the prediction variable is called .pred_class.
predict(object = iris_ranger, new_data = iris_testing_baked) %>%
dplyr::bind_cols(iris_testing_baked) %>%
glimpse()Rows: 60
Columns: 5
$ .pred_class <fct> setosa, setosa, setosa, setosa, setosa, setosa, setosa, s…
$ Sepal.Length <dbl> -1.5852786, -0.5918925, -1.8336251, -1.3369321, -1.336932…
$ Sepal.Width <dbl> 0.05284097, 1.78218168, -0.37949421, 0.70134373, -0.16332…
$ Petal.Width <dbl> -1.3124100, -1.0448745, -1.3124100, -1.3124100, -1.446177…
$ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, setosa, s…Iris Classification Model Validation
We use metrics() function from the yardstick package to evaluate how good the model is.
predict(iris_ranger, iris_testing_baked) %>%
dplyr::bind_cols(iris_testing_baked) %>%
yardstick::metrics(truth = Species, estimate = .pred_class)We can also check the metrics for randomForest model:
Iris Per-Classifier Metrics
We can use the parameter type = "prob" in the predict() function to obtain a probability score on each prediction. TBD: How is this prob calculated? Possible answer: the Random Forest model outputs its answer by majority voting across n trees. Each of the possible answers( i.e. predictions) for a particular test datum gets a share of the vote, that represents its probability. Hence each dataum in the test vector can show a probability for the “winning” answer. ( Quite possibly we can get the probabilities for all possible outcomes for each test datum)
iris_ranger_probs <-
predict(iris_ranger, iris_testing_baked, type = "prob") %>%
dplyr::bind_cols(iris_testing_baked)
glimpse(iris_ranger_probs)Rows: 60
Columns: 7
$ .pred_setosa <dbl> 0.980329365, 0.980809524, 0.887333333, 0.964476190, 0…
$ .pred_versicolor <dbl> 0.01967063, 0.00900000, 0.10541667, 0.02385714, 0.014…
$ .pred_virginica <dbl> 0.000000000, 0.010190476, 0.007250000, 0.011666667, 0…
$ Sepal.Length <dbl> -1.5852786, -0.5918925, -1.8336251, -1.3369321, -1.33…
$ Sepal.Width <dbl> 0.05284097, 1.78218168, -0.37949421, 0.70134373, -0.1…
$ Petal.Width <dbl> -1.3124100, -1.0448745, -1.3124100, -1.3124100, -1.44…
$ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, setos…iris_rf_probs <-
predict(iris_rf, iris_testing_baked, type = "prob") %>%
dplyr::bind_cols(iris_testing_baked)
glimpse(iris_rf_probs)Rows: 60
Columns: 7
$ .pred_setosa <dbl> 1.00, 1.00, 0.94, 0.99, 1.00, 0.87, 0.99, 0.99, 0.99,…
$ .pred_versicolor <dbl> 0.00, 0.00, 0.06, 0.00, 0.00, 0.10, 0.00, 0.00, 0.00,…
$ .pred_virginica <dbl> 0.00, 0.00, 0.00, 0.01, 0.00, 0.03, 0.01, 0.01, 0.01,…
$ Sepal.Length <dbl> -1.5852786, -0.5918925, -1.8336251, -1.3369321, -1.33…
$ Sepal.Width <dbl> 0.05284097, 1.78218168, -0.37949421, 0.70134373, -0.1…
$ Petal.Width <dbl> -1.3124100, -1.0448745, -1.3124100, -1.3124100, -1.44…
$ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, setos…# Tabulating the probabilities
ftable(iris_rf_probs$.pred_versicolor) 0 0.01 0.02 0.03 0.04 0.05 0.06 0.08 0.1 0.18 0.2 0.24 0.27 0.3 0.33 0.43 0.59 0.61 0.63 0.7 0.71 0.8 0.83 0.84 0.86 0.9 0.91 0.93 0.94 1
16 3 1 4 1 1 4 2 3 1 1 1 1 2 1 1 2 1 1 1 1 1 2 1 1 1 1 2 1 1ftable(iris_rf_probs$.pred_virginica) 0 0.01 0.03 0.05 0.06 0.07 0.08 0.09 0.14 0.16 0.17 0.19 0.29 0.37 0.41 0.57 0.67 0.69 0.7 0.73 0.75 0.79 0.82 0.9 0.92 0.94 0.95 0.97 0.99 1
14 7 1 2 1 2 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 2 2 5 1 1 3 1ftable(iris_rf_probs$.pred_setosa) 0 0.01 0.03 0.04 0.05 0.1 0.2 0.24 0.87 0.94 0.95 0.96 0.98 0.99 1
23 9 2 1 1 1 2 1 1 2 1 1 1 5 9Iris Classifier: Gain and ROC Curves
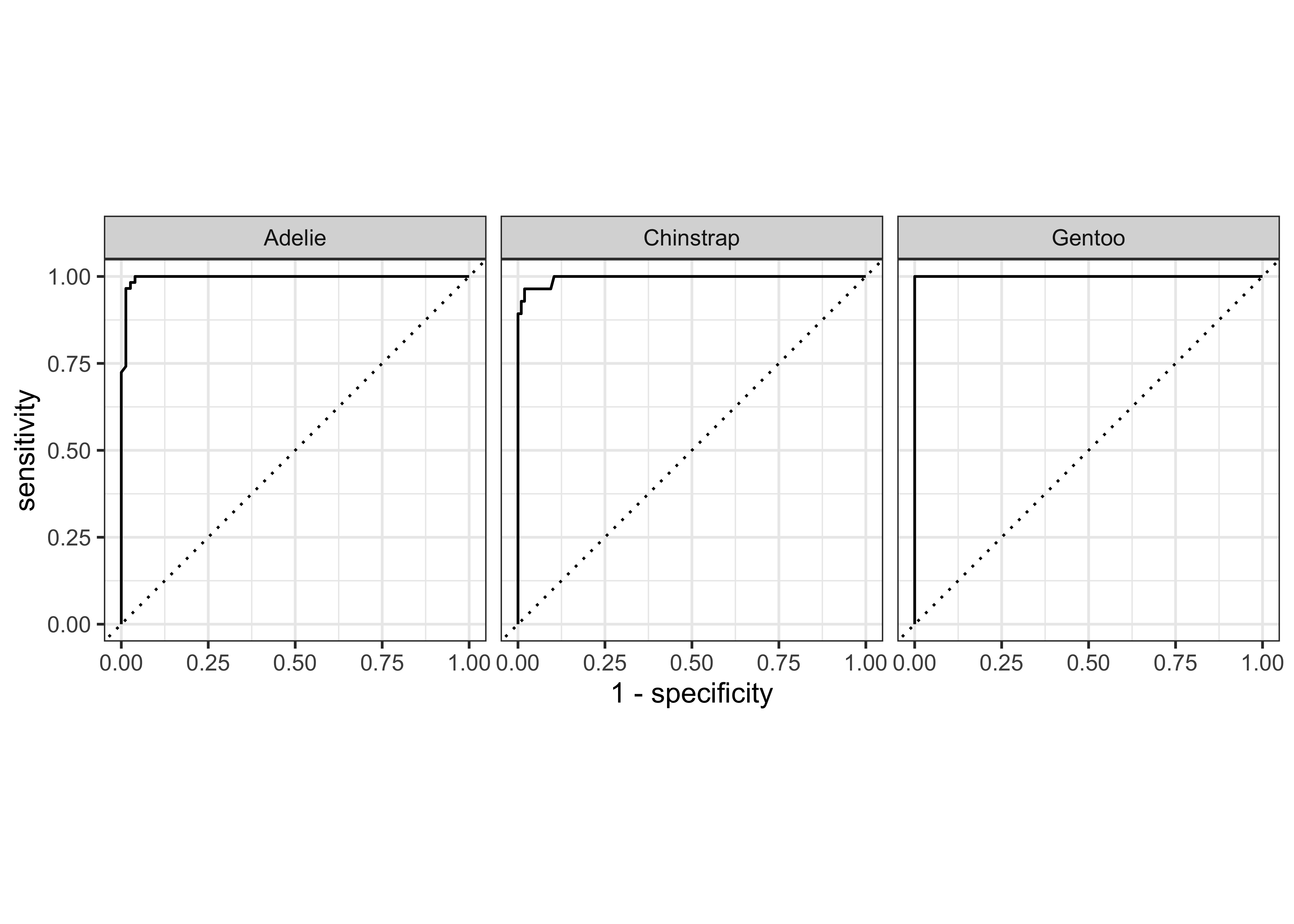
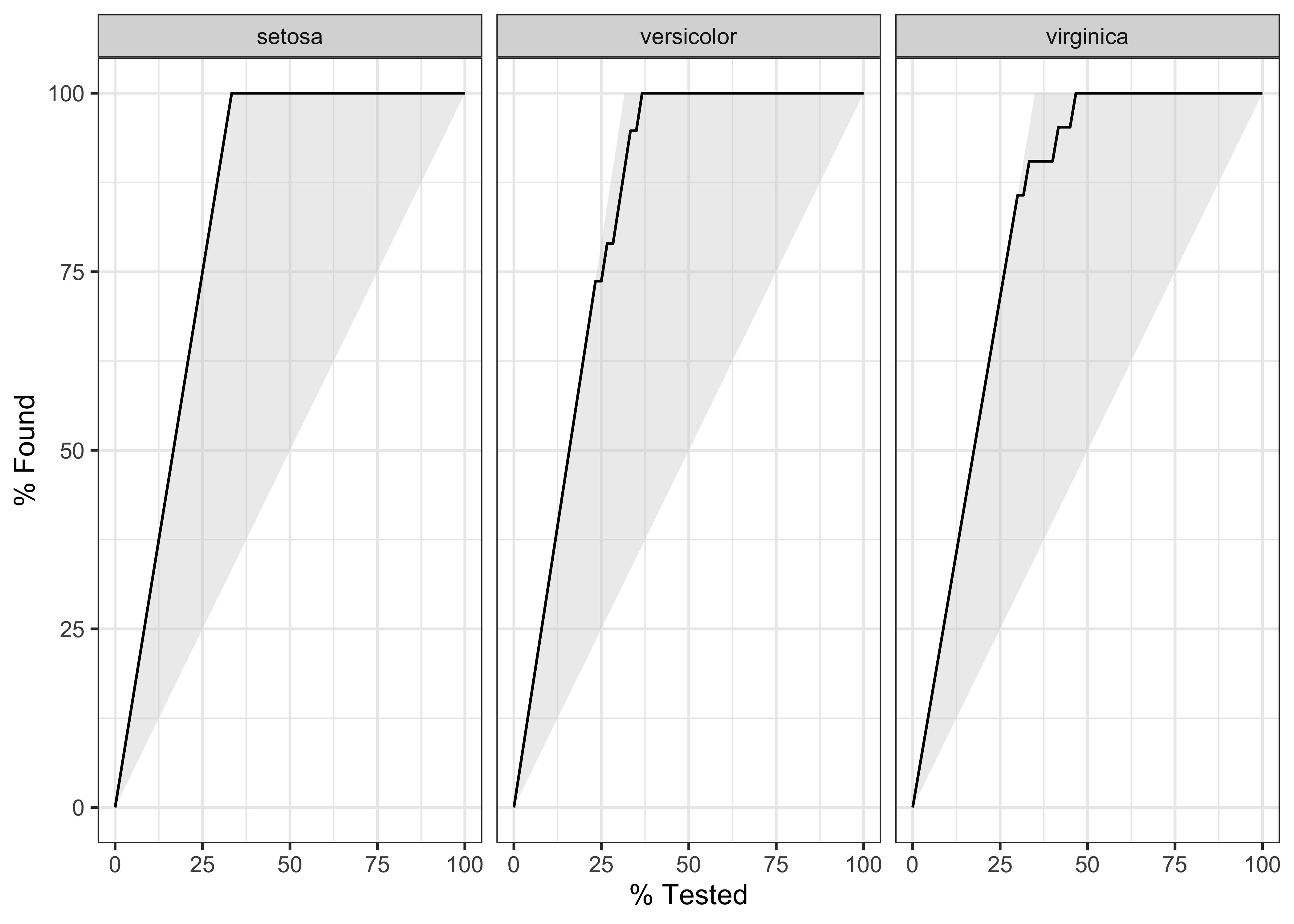
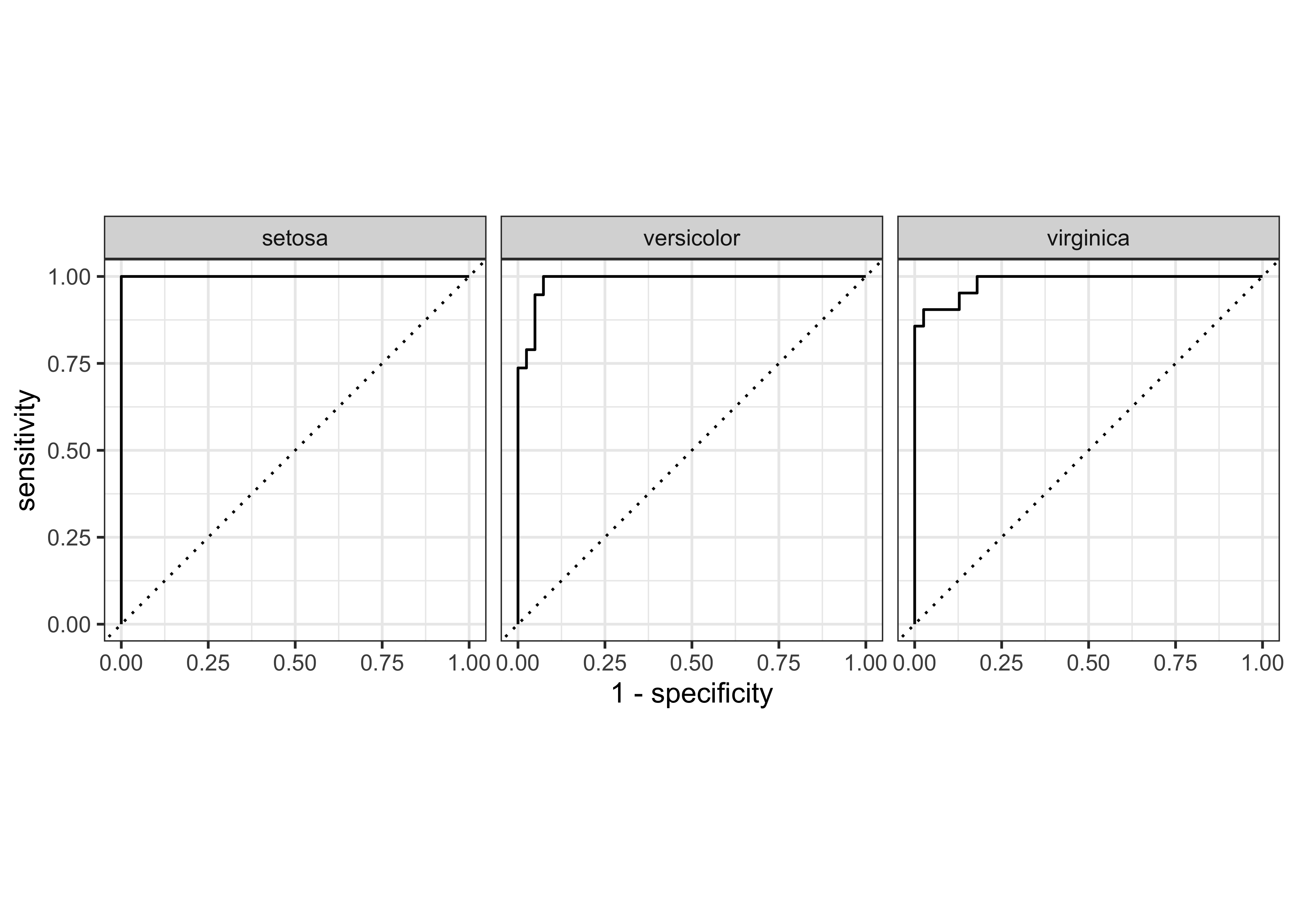
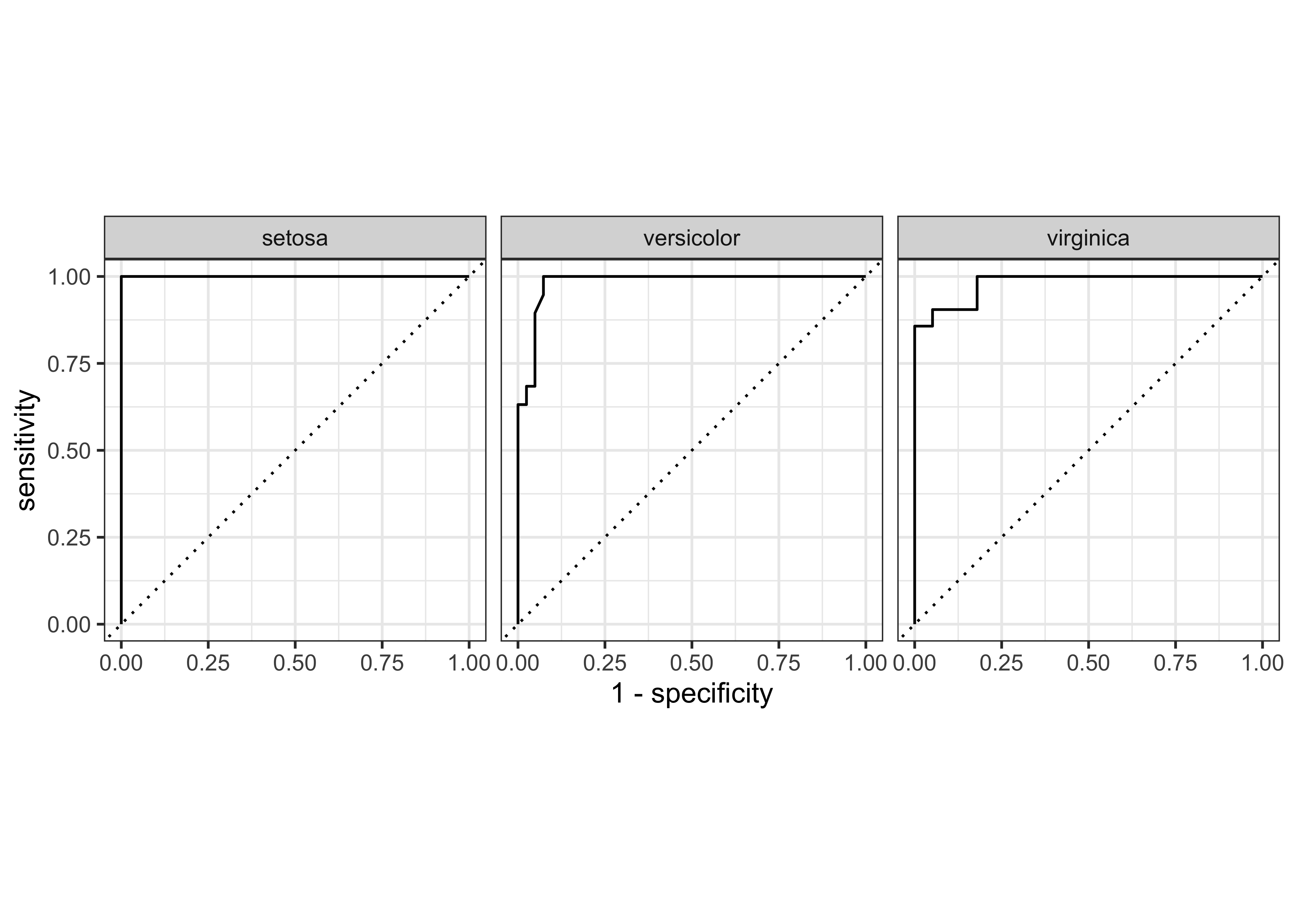
We can plot gain and ROC curves for each of these models
iris_ranger_probs %>%
yardstick::gain_curve(Species, .pred_setosa:.pred_virginica) %>%
glimpse()Rows: 145
Columns: 5
$ .level <chr> "setosa", "setosa", "setosa", "setosa", "setosa", "set…
$ .n <dbl> 0, 1, 3, 4, 6, 7, 9, 10, 12, 13, 14, 15, 16, 17, 18, 1…
$ .n_events <dbl> 0, 1, 3, 4, 6, 7, 9, 10, 12, 13, 14, 15, 16, 17, 18, 1…
$ .percent_tested <dbl> 0.000000, 1.666667, 5.000000, 6.666667, 10.000000, 11.…
$ .percent_found <dbl> 0, 5, 15, 20, 30, 35, 45, 50, 60, 65, 70, 75, 80, 85, …iris_ranger_probs %>%
yardstick::gain_curve(Species, .pred_setosa:.pred_virginica) %>%
autoplot()Rows: 148
Columns: 4
$ .level <chr> "setosa", "setosa", "setosa", "setosa", "setosa", "setosa"…
$ .threshold <dbl> -Inf, 0.000000000, 0.001111111, 0.002000000, 0.002361111, …
$ specificity <dbl> 0.000, 0.000, 0.225, 0.275, 0.300, 0.325, 0.375, 0.500, 0.…
$ sensitivity <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…iris_rf_probs %>%
yardstick::gain_curve(Species, .pred_setosa:.pred_virginica) %>%
glimpse()Rows: 78
Columns: 5
$ .level <chr> "setosa", "setosa", "setosa", "setosa", "setosa", "set…
$ .n <dbl> 0, 9, 14, 15, 16, 17, 19, 20, 21, 23, 24, 25, 26, 28, …
$ .n_events <dbl> 0, 9, 14, 15, 16, 17, 19, 20, 20, 20, 20, 20, 20, 20, …
$ .percent_tested <dbl> 0.000000, 15.000000, 23.333333, 25.000000, 26.666667, …
$ .percent_found <dbl> 0.000000, 45.000000, 70.000000, 75.000000, 80.000000, …iris_rf_probs %>%
yardstick::gain_curve(Species, .pred_setosa:.pred_virginica) %>%
autoplot()Rows: 81
Columns: 4
$ .level <chr> "setosa", "setosa", "setosa", "setosa", "setosa", "setosa"…
$ .threshold <dbl> -Inf, 0.00, 0.01, 0.03, 0.04, 0.05, 0.10, 0.20, 0.24, 0.87…
$ specificity <dbl> 0.0000000, 0.0000000, 0.5750000, 0.8000000, 0.8500000, 0.8…
$ sensitivity <dbl> 1.00, 1.00, 1.00, 1.00, 1.00, 1.00, 1.00, 1.00, 1.00, 1.00…Iris Classifier: Metrics
predict(iris_ranger, iris_testing_baked, type = "prob") %>%
bind_cols(predict(iris_ranger, iris_testing_baked)) %>%
bind_cols(select(iris_testing_baked, Species)) %>%
glimpse()Rows: 60
Columns: 5
$ .pred_setosa <dbl> 0.980329365, 0.980809524, 0.887333333, 0.964476190, 0…
$ .pred_versicolor <dbl> 0.01967063, 0.00900000, 0.10541667, 0.02385714, 0.014…
$ .pred_virginica <dbl> 0.000000000, 0.010190476, 0.007250000, 0.011666667, 0…
$ .pred_class <fct> setosa, setosa, setosa, setosa, setosa, setosa, setos…
$ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, setos…# predict(iris_ranger, iris_testing_baked, type = "prob") %>%
# bind_cols(predict(iris_ranger,iris_testing_baked)) %>%
# bind_cols(select(iris_testing_baked,Species)) %>%
# yardstick::metrics(data = ., truth = Species, estimate = .pred_class, ... = .pred_setosa:.pred_virginica)
# And for the `randomForest`method
predict(iris_rf, iris_testing_baked, type = "prob") %>%
bind_cols(predict(iris_ranger, iris_testing_baked)) %>%
bind_cols(select(iris_testing_baked, Species)) %>%
glimpse()Rows: 60
Columns: 5
$ .pred_setosa <dbl> 1.00, 1.00, 0.94, 0.99, 1.00, 0.87, 0.99, 0.99, 0.99,…
$ .pred_versicolor <dbl> 0.00, 0.00, 0.06, 0.00, 0.00, 0.10, 0.00, 0.00, 0.00,…
$ .pred_virginica <dbl> 0.00, 0.00, 0.00, 0.01, 0.00, 0.03, 0.01, 0.01, 0.01,…
$ .pred_class <fct> setosa, setosa, setosa, setosa, setosa, setosa, setos…
$ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, setos…# predict(iris_rf, iris_testing_baked, type = "prob") %>%
# bind_cols(predict(iris_ranger,iris_testing_baked)) %>%
# bind_cols(select(iris_testing_baked,Species)) %>%
# yardstick::metrics(data = ., truth = Species, estimate = .pred_class, ... = .pred_setosa:.pred_virginica)References
- Machine Learning Basics - Random Forest at Shirin’s Playground