Tutorial: Multiple Linear Regression with Backward Selection
# Let us set a plot theme for Data visualisation
# my_theme <- function(){ # Creating a function
# theme_classic() + # Using pre-defined theme as base
# theme(axis.text.x = element_text(size = 12, face = "bold"), # Customizing axes text
# axis.text.y = element_text(size = 12, face = "bold"),
# axis.title = element_text(size = 14, face = "bold"), # Customizing axis title
# panel.grid = element_blank(), # Taking off the default grid
# plot.margin = unit(c(0.5, 0.5, 0.5, 0.5), units = , "cm"),
# legend.text = element_text(size = 12, face = "italic"), # Customizing legend text
# legend.title = element_text(size = 12, face = "bold"), # Customizing legend title
# legend.position = "right", # Customizing legend position
# plot.caption = element_text(size = 12)) # Customizing plot caption
# }
my_theme <- function() { # Creating a function
theme_classic() + # Using pre-defined theme as base
theme(
plot.title = element_text(face = "bold", size = 14),
axis.text.x = element_text(size = 10, face = "bold"),
# Customizing axes text
axis.text.y = element_text(size = 10, face = "bold"),
axis.title = element_text(size = 12, face = "bold"),
# Customizing axis title
panel.grid = element_blank(), # Taking off the default grid
plot.margin = unit(c(0.5, 0.5, 0.5, 0.5), units = , "cm"),
legend.text = element_text(size = 8, face = "italic"),
# Customizing legend text
legend.title = element_text(size = 10, face = "bold"),
# Customizing legend title
legend.position = "right", # Customizing legend position
plot.caption = element_text(size = 8)
) # Customizing plot caption
}In this tutorial, we will use the Boston housing dataset. Our research question is:
How do we predict the price of a house in Boston, based on other parameters Quantitative parameters such as area, location, rooms, and crime-rate in the neighbourhood?
And how do we choose the “best” model, based on a tradeoff between Model Complexity and Model Accuracy?
categorical variables:
name class levels n missing distribution
1 town factor 92 506 0 Cambridge (5.9%) ...
2 chas factor 2 506 0 0 (93.1%), 1 (6.9%)
quantitative variables:
name class min Q1 median Q3 max mean
1 tract integer 1.00000 1303.250 3393.500 3739.750 5082.000 2700.356
2 lon numeric -71.28950 -71.093 -71.053 -71.020 -70.810 -71.056
3 lat numeric 42.03000 42.181 42.218 42.252 42.381 42.216
4 medv numeric 5.00000 17.025 21.200 25.000 50.000 22.533
5 cmedv numeric 5.00000 17.025 21.200 25.000 50.000 22.529
6 crim numeric 0.00632 0.082 0.257 3.677 88.976 3.614
7 zn numeric 0.00000 0.000 0.000 12.500 100.000 11.364
8 indus numeric 0.46000 5.190 9.690 18.100 27.740 11.137
9 nox numeric 0.38500 0.449 0.538 0.624 0.871 0.555
10 rm numeric 3.56100 5.886 6.208 6.623 8.780 6.285
11 age numeric 2.90000 45.025 77.500 94.075 100.000 68.575
12 dis numeric 1.12960 2.100 3.207 5.188 12.127 3.795
13 rad integer 1.00000 4.000 5.000 24.000 24.000 9.549
14 tax integer 187.00000 279.000 330.000 666.000 711.000 408.237
15 ptratio numeric 12.60000 17.400 19.050 20.200 22.000 18.456
16 b numeric 0.32000 375.377 391.440 396.225 396.900 356.674
17 lstat numeric 1.73000 6.950 11.360 16.955 37.970 12.653
sd n missing
1 1380.0368 506 0
2 0.0754 506 0
3 0.0618 506 0
4 9.1971 506 0
5 9.1822 506 0
6 8.6015 506 0
7 23.3225 506 0
8 6.8604 506 0
9 0.1159 506 0
10 0.7026 506 0
11 28.1489 506 0
12 2.1057 506 0
13 8.7073 506 0
14 168.5371 506 0
15 2.1649 506 0
16 91.2949 506 0
17 7.1411 506 0The original data are 506 observations on 14 variables, medv being the target variable:
| crim | per capita crime rate by town |
| zn | proportion of residential land zoned for lots over 25,000 sq.ft |
| indus | proportion of non-retail business acres per town |
| chas | Charles River dummy variable (= 1 if tract bounds river; 0 otherwise) |
| nox | nitric oxides concentration (parts per 10 million) |
| rm | average number of rooms per dwelling |
| age | proportion of owner-occupied units built prior to 1940 |
| dis | weighted distances to five Boston employment centres |
| rad | index of accessibility to radial highways |
| tax | full-value property-tax rate per USD 10,000 |
| ptratio | pupil-teacher ratio by town |
| b |
|
| lstat | percentage of lower status of the population |
| medv | median value of owner-occupied homes in USD 1000’s |
The corrected data set has the following additional columns:
| cmedv | corrected median value of owner-occupied homes in USD 1000’s |
| town | name of town |
| tract | census tract |
| lon | longitude of census tract |
| lat | latitude of census tract |
Our response variable is cmedv, the corrected median value of owner-occupied homes in USD 1000’s. Their are many Quantitative feature variables that we can use to predict cmedv. And there are two Qualitative features, chas and tax.
We can use purrr to evaluate all pair-wise correlations in one shot:
all_corrs <- housing %>%
select(where(is.numeric)) %>%
# leave off cmedv/medv to get all the remaining ones
select(-cmedv, -medv) %>%
# perform a cor.test for all variables against cmedv
purrr::map(
.x = .,
.f = \(x) cor.test(x, housing$cmedv)
) %>%
# tidy up the cor.test outputs into a tidy data frame
map_dfr(broom::tidy, .id = "predictor")
all_corrspredictor <chr> | estimate <dbl> | statistic <dbl> | p.value <dbl> | parameter <int> | conf.low <dbl> | conf.high <dbl> | method <chr> | |
|---|---|---|---|---|---|---|---|---|
| tract | 0.42825 | 10.639 | 5.51e-24 | 504 | 0.3543 | 0.4969 | Pearson's product-moment correlation | |
| lon | -0.32295 | -7.661 | 9.55e-14 | 504 | -0.3989 | -0.2426 | Pearson's product-moment correlation | |
| lat | 0.00683 | 0.153 | 8.78e-01 | 504 | -0.0804 | 0.0939 | Pearson's product-moment correlation | |
| crim | -0.38958 | -9.496 | 8.71e-20 | 504 | -0.4611 | -0.3130 | Pearson's product-moment correlation | |
| zn | 0.36039 | 8.673 | 5.79e-17 | 504 | 0.2821 | 0.4339 | Pearson's product-moment correlation | |
| indus | -0.48475 | -12.442 | 3.52e-31 | 504 | -0.5487 | -0.4151 | Pearson's product-moment correlation | |
| nox | -0.42930 | -10.671 | 4.17e-24 | 504 | -0.4978 | -0.3554 | Pearson's product-moment correlation | |
| rm | 0.69630 | 21.779 | 1.31e-74 | 504 | 0.6485 | 0.7386 | Pearson's product-moment correlation | |
| age | -0.37800 | -9.166 | 1.24e-18 | 504 | -0.4503 | -0.3007 | Pearson's product-moment correlation | |
| dis | 0.24931 | 5.780 | 1.31e-08 | 504 | 0.1657 | 0.3293 | Pearson's product-moment correlation |
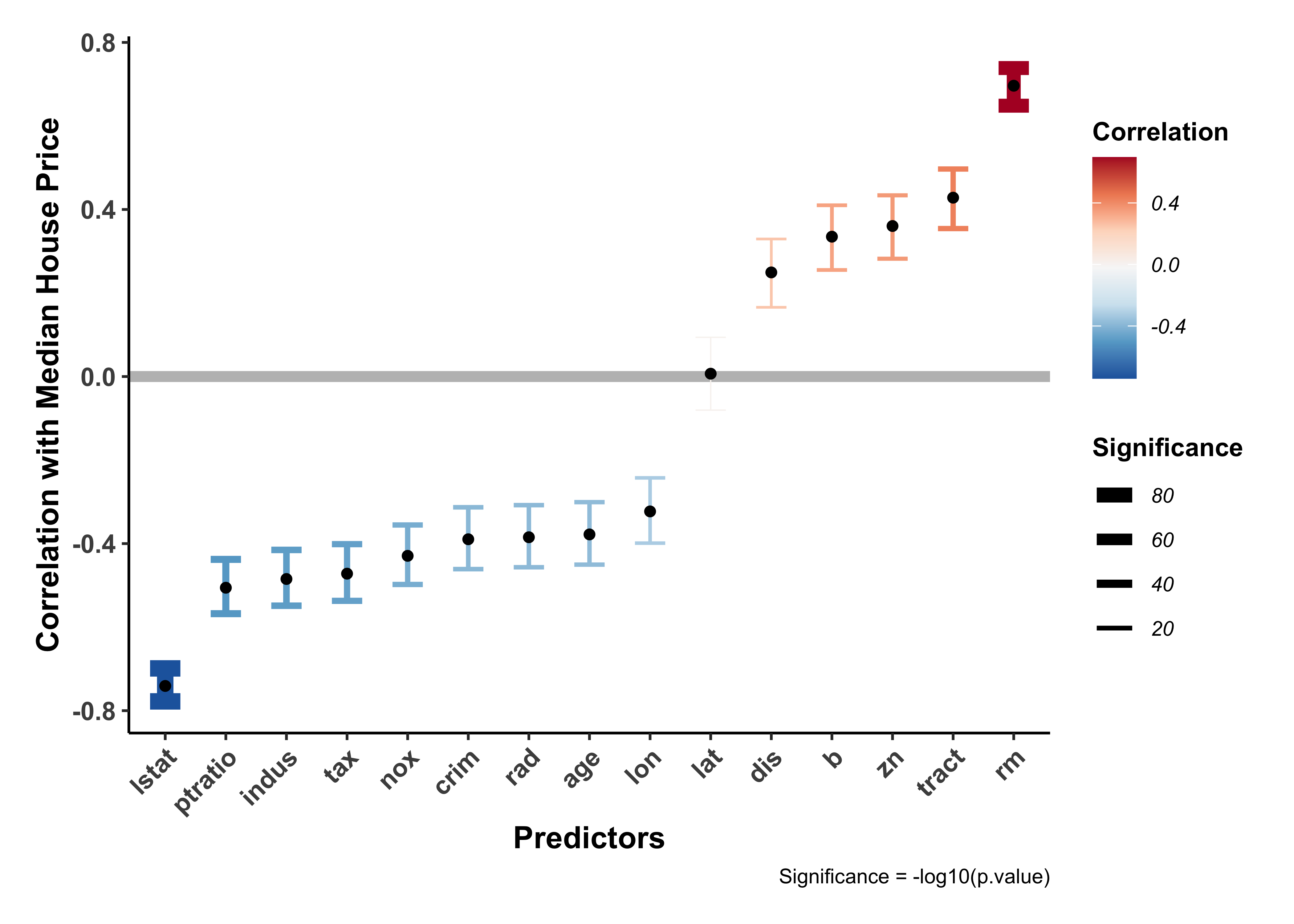
all_corrs %>%
gf_hline(
yintercept = 0,
color = "grey",
linewidth = 2
) %>%
gf_errorbar(
conf.high + conf.low ~ reorder(predictor, estimate),
colour = ~estimate,
width = 0.5,
linewidth = ~ -log10(p.value),
caption = "Significance = -log10(p.value)"
) %>%
gf_point(estimate ~ reorder(predictor, estimate)) %>%
gf_labs(x = "Predictors", y = "Correlation with Median House Price") %>%
gf_theme(my_theme()) %>%
gf_theme(theme(axis.text.x = element_text(angle = 45, hjust = 1))) %>%
gf_refine(
scale_colour_distiller("Correlation", type = "div", palette = "RdBu"),
scale_linewidth_continuous("Significance", range = c(0.25, 3)),
guides(linewidth = guide_legend(reverse = TRUE))
)The variables rm, lstat seem to have high correlations with cmedv which are also statistically significant.
We will create a regression model for cmedv using all the other numerical predictor features in the dataset.
housing_numeric <- housing %>% select(
where(is.numeric),
# remove medv
# an older version of cmedv
-c(medv)
)
names(housing_numeric) # 16 variables, one target, 15 predictors [1] "tract" "lon" "lat" "cmedv" "crim" "zn" "indus"
[8] "nox" "rm" "age" "dis" "rad" "tax" "ptratio"
[15] "b" "lstat"
Call:
lm(formula = cmedv ~ ., data = housing_numeric)
Residuals:
Min 1Q Median 3Q Max
-13.934 -2.752 -0.619 1.711 26.120
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3.45e+02 3.23e+02 -1.07 0.28734
tract -7.52e-04 4.46e-04 -1.69 0.09231 .
lon -6.79e+00 3.44e+00 -1.98 0.04870 *
lat -2.35e+00 5.36e+00 -0.44 0.66074
crim -1.09e-01 3.28e-02 -3.32 0.00097 ***
zn 4.40e-02 1.39e-02 3.17 0.00164 **
indus 2.75e-02 6.20e-02 0.44 0.65692
nox -1.55e+01 4.03e+00 -3.85 0.00014 ***
rm 3.81e+00 4.20e-01 9.07 < 2e-16 ***
age 5.82e-03 1.34e-02 0.43 0.66416
dis -1.38e+00 2.10e-01 -6.59 1.1e-10 ***
rad 2.36e-01 8.47e-02 2.78 0.00558 **
tax -1.48e-02 3.74e-03 -3.96 8.5e-05 ***
ptratio -9.49e-01 1.41e-01 -6.73 4.7e-11 ***
b 9.55e-03 2.67e-03 3.57 0.00039 ***
lstat -5.46e-01 5.06e-02 -10.80 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 4.73 on 490 degrees of freedom
Multiple R-squared: 0.743, Adjusted R-squared: 0.735
F-statistic: 94.3 on 15 and 490 DF, p-value: <2e-16The maximal model has an R.squared of rm alone. How much can we simplify this maximal model, without losing out on R.squared?
We now proceed naively by removing one predictor after another. We will resort to what may amount to p-hacking by sorting the predictors based on their p-value1 in the maximal model and removing them in decreasing order of their p-value.
We will also use some powerful features from the purrr package (also part of the tidyverse), to create all these models all at once. Then we will be able to plot their R.squared values together and decide where we wish to trade off Explainability vs Complexity for our model.
# No of Quant predictor variables in the dataset
n_vars <- housing %>%
select(where(is.numeric), -c(cmedv, medv)) %>%
length()
# Maximal Model, now tidied
housing_maximal_tidy <-
housing_maximal %>%
broom::tidy() %>%
# Obviously remove "Intercept" ;-D
filter(term != "(Intercept)") %>%
# And horrors! Sort variables by p.value
arrange(p.value)
housing_maximal_tidyterm <chr> | estimate <dbl> | std.error <dbl> | statistic <dbl> | p.value <dbl> |
|---|---|---|---|---|
| lstat | -0.546440 | 0.050619 | -10.795 | 1.62e-24 |
| rm | 3.809475 | 0.419779 | 9.075 | 2.77e-18 |
| ptratio | -0.948941 | 0.140991 | -6.730 | 4.74e-11 |
| dis | -1.384370 | 0.210041 | -6.591 | 1.13e-10 |
| tax | -0.014807 | 0.003737 | -3.962 | 8.54e-05 |
| nox | -15.506379 | 4.032786 | -3.845 | 1.36e-04 |
| b | 0.009548 | 0.002675 | 3.569 | 3.94e-04 |
| crim | -0.108861 | 0.032789 | -3.320 | 9.67e-04 |
| zn | 0.043968 | 0.013883 | 3.167 | 1.64e-03 |
| rad | 0.235674 | 0.084665 | 2.784 | 5.58e-03 |
The last 5 variables are clearly statistically insignificant.
And now we unleash the purrr package to create all the simplified models at once. We will construct a dataset containing three columns:
- A list of all quantitative predictor variables
- A sequence of numbers from 1 to
N(predictor) - A “list” column containing the
housingdata frame itself
We will use the iteration capability of purrr to sequentially drop one variable at a time from the maximal(15 predictor) model, build a new reduced model each time, and compute the r.squared:
housing_model_set <- tibble(
all_vars =
list(housing_maximal_tidy$term), # p-hacked order!!
keep_vars = seq(1, n_vars),
data = list(housing_numeric)
)
housing_model_set
# Unleash purrr in a series of mutates
housing_model_set <- housing_model_set %>%
# list of predictor variables for each model
mutate(
mod_vars =
pmap(
.l = list(all_vars, keep_vars, data),
.f = \(all_vars, keep_vars, data) all_vars[1:keep_vars]
)
) %>%
# build formulae with these for linear regression
mutate(
formula =
map(
.x = mod_vars,
.f = \(mod_vars) as.formula(paste(
"cmedv ~", paste(mod_vars, collapse = "+")
))
)
) %>%
# use the formulae to build multiple linear models
mutate(
models =
pmap(
.l = list(data, formula),
.f = \(data, formula) lm(formula, data = data)
)
)
# Check everything after the operation
housing_model_set
# Tidy up the models using broom to expose their metrics
housing_models_tidy <- housing_model_set %>%
mutate(
tidy_models =
map(
.x = models,
.f = \(x) broom::glance(x,
conf.int = TRUE,
conf.lvel = 0.95
)
)
) %>%
# Remove unwanted columns, keep model and predictor count
select(keep_vars, tidy_models) %>%
unnest(tidy_models)
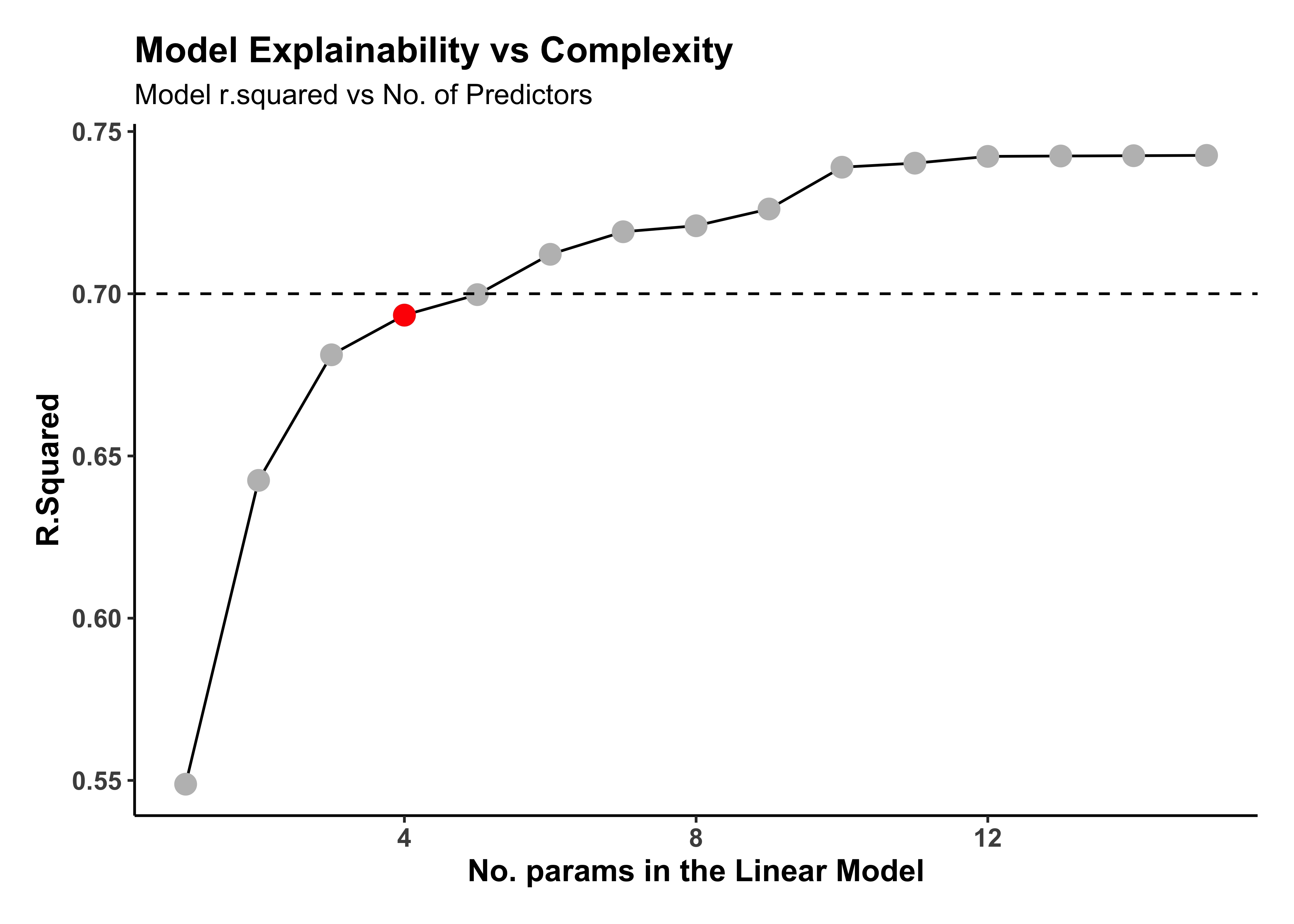
housing_models_tidy %>%
gf_line(
r.squared ~ keep_vars,
ylab = "R.Squared",
xlab = "No. params in the Linear Model",
title = "Model Explainability vs Complexity",
subtitle = "Model r.squared vs No. of Predictors",
data = .
) %>%
# Plot r.squared vs predictor count
gf_point(r.squared ~ keep_vars,
size = 3.5, color = "grey"
) %>%
# Show off the selected best model
gf_point(
r.squared ~ keep_vars,
size = 3.5,
color = "red",
data = housing_models_tidy %>% filter(keep_vars == 4)
) %>%
gf_hline(yintercept = 0.7, linetype = "dashed") %>%
gf_theme(my_theme())all_vars <list> | keep_vars <int> | data <list> | |
|---|---|---|---|
| <chr [15]> | 1 | <df[,16] [506 × 16]> | |
| <chr [15]> | 2 | <df[,16] [506 × 16]> | |
| <chr [15]> | 3 | <df[,16] [506 × 16]> | |
| <chr [15]> | 4 | <df[,16] [506 × 16]> | |
| <chr [15]> | 5 | <df[,16] [506 × 16]> | |
| <chr [15]> | 6 | <df[,16] [506 × 16]> | |
| <chr [15]> | 7 | <df[,16] [506 × 16]> | |
| <chr [15]> | 8 | <df[,16] [506 × 16]> | |
| <chr [15]> | 9 | <df[,16] [506 × 16]> | |
| <chr [15]> | 10 | <df[,16] [506 × 16]> |
all_vars <list> | keep_vars <int> | data <list> | mod_vars <list> | |
|---|---|---|---|---|
| <chr [15]> | 1 | <df[,16] [506 × 16]> | <chr [1]> | |
| <chr [15]> | 2 | <df[,16] [506 × 16]> | <chr [2]> | |
| <chr [15]> | 3 | <df[,16] [506 × 16]> | <chr [3]> | |
| <chr [15]> | 4 | <df[,16] [506 × 16]> | <chr [4]> | |
| <chr [15]> | 5 | <df[,16] [506 × 16]> | <chr [5]> | |
| <chr [15]> | 6 | <df[,16] [506 × 16]> | <chr [6]> | |
| <chr [15]> | 7 | <df[,16] [506 × 16]> | <chr [7]> | |
| <chr [15]> | 8 | <df[,16] [506 × 16]> | <chr [8]> | |
| <chr [15]> | 9 | <df[,16] [506 × 16]> | <chr [9]> | |
| <chr [15]> | 10 | <df[,16] [506 × 16]> | <chr [10]> |
At the loss of some 5% in the r.squared, we can drop our model complexity from 15 predictors to say 4! Our final model will then be:
housing_model_final <-
housing_model_set %>%
# filter for best model, with 4 variables
filter(keep_vars == 4) %>%
# tidy up the model
mutate(
tidy_models =
map(
.x = models,
.f = \(x) broom::tidy(x,
conf.int = TRUE,
conf.lvel = 0.95
)
)
) %>%
# Remove unwanted columns, keep model and predictor count
select(keep_vars, models, tidy_models) %>%
unnest(tidy_models)
housing_model_finalkeep_vars <int> | models <list> | term <chr> | estimate <dbl> | std.error <dbl> | statistic <dbl> | p.value <dbl> | conf.low <dbl> | conf.high <dbl> |
|---|---|---|---|---|---|---|---|---|
| 4 | <S3: lm> | (Intercept) | 24.459 | 4.0510 | 6.04 | 3.04e-09 | 16.500 | 32.418 |
| 4 | <S3: lm> | lstat | -0.673 | 0.0464 | -14.49 | 5.36e-40 | -0.764 | -0.582 |
| 4 | <S3: lm> | rm | 4.199 | 0.4210 | 9.97 | 1.71e-21 | 3.372 | 5.026 |
| 4 | <S3: lm> | ptratio | -0.957 | 0.1153 | -8.30 | 9.41e-16 | -1.184 | -0.731 |
| 4 | <S3: lm> | dis | -0.564 | 0.1261 | -4.47 | 9.74e-06 | -0.811 | -0.316 |
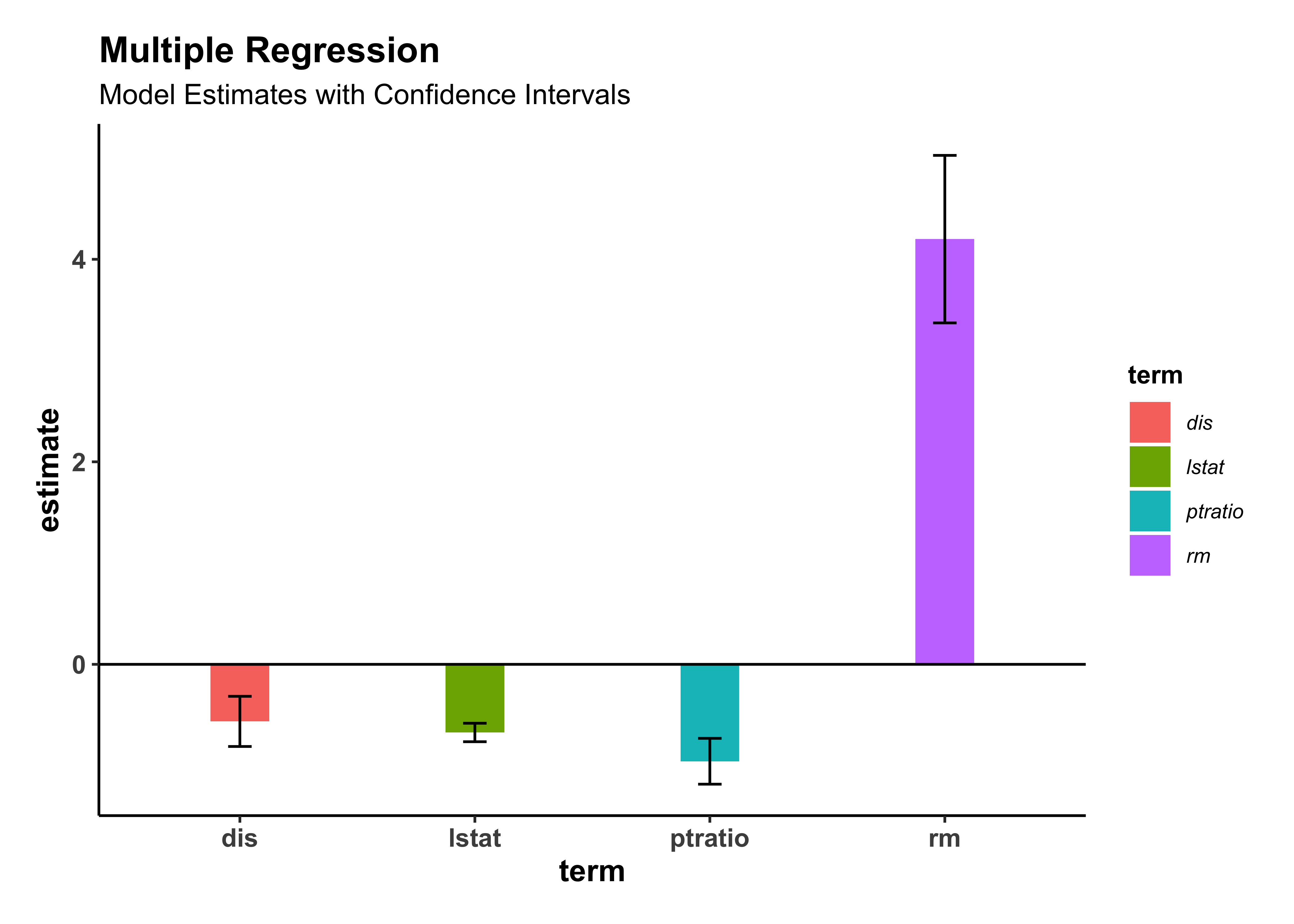
housing_model_final %>%
# And plot the model
# Remove the intercept term
filter(term != "(Intercept)") %>%
gf_col(estimate ~ term, fill = ~term, width = 0.25) %>%
gf_hline(yintercept = 0) %>%
gf_errorbar(conf.low + conf.high ~ term,
width = 0.1,
title = "Multiple Regression",
subtitle = "Model Estimates with Confidence Intervals"
) %>%
gf_theme(my_theme())Our current best model can be stated as:
Let us use broom::augment to calculate residuals and predictions to arrive at a quick set of diagnostic plots.
housing_model_final_augment <-
housing_model_set %>%
filter(keep_vars == 4) %>%
# augment the model
mutate(
augment_models =
map(
.x = models,
.f = \(x) broom::augment(x)
)
) %>%
unnest(augment_models) %>%
select(cmedv:last_col())
housing_model_final_augmentcmedv <dbl> | lstat <dbl> | rm <dbl> | ptratio <dbl> | dis <dbl> | .fitted <dbl> | .resid <dbl> | .hat <dbl> | .sigma <dbl> | .cooksd <dbl> | |
|---|---|---|---|---|---|---|---|---|---|---|
| 24.0 | 4.98 | 6.58 | 15.3 | 4.09 | 31.767 | -7.7666 | 0.00809 | 5.10 | 3.81e-03 | |
| 21.6 | 9.14 | 6.42 | 17.8 | 4.97 | 25.433 | -3.8328 | 0.00276 | 5.11 | 3.13e-04 | |
| 34.7 | 4.03 | 7.18 | 17.8 | 4.97 | 32.080 | 2.6198 | 0.00596 | 5.11 | 3.18e-04 | |
| 33.4 | 2.94 | 7.00 | 18.7 | 6.06 | 30.550 | 2.8501 | 0.00740 | 5.11 | 4.68e-04 | |
| 36.2 | 5.33 | 7.15 | 18.7 | 6.06 | 29.567 | 6.6330 | 0.00747 | 5.10 | 2.56e-03 | |
| 28.7 | 5.21 | 6.43 | 18.7 | 6.06 | 26.637 | 2.0630 | 0.00587 | 5.11 | 1.94e-04 | |
| 22.9 | 12.43 | 6.01 | 15.2 | 5.56 | 23.655 | -0.7554 | 0.00947 | 5.11 | 4.23e-05 | |
| 22.1 | 19.15 | 6.17 | 15.2 | 5.95 | 19.585 | 2.5153 | 0.01679 | 5.11 | 8.44e-04 | |
| 16.5 | 29.93 | 5.63 | 15.2 | 6.08 | 9.984 | 6.5165 | 0.04008 | 5.10 | 1.42e-02 | |
| 18.9 | 17.10 | 6.00 | 15.2 | 6.59 | 19.897 | -0.9974 | 0.01599 | 5.11 | 1.26e-04 |
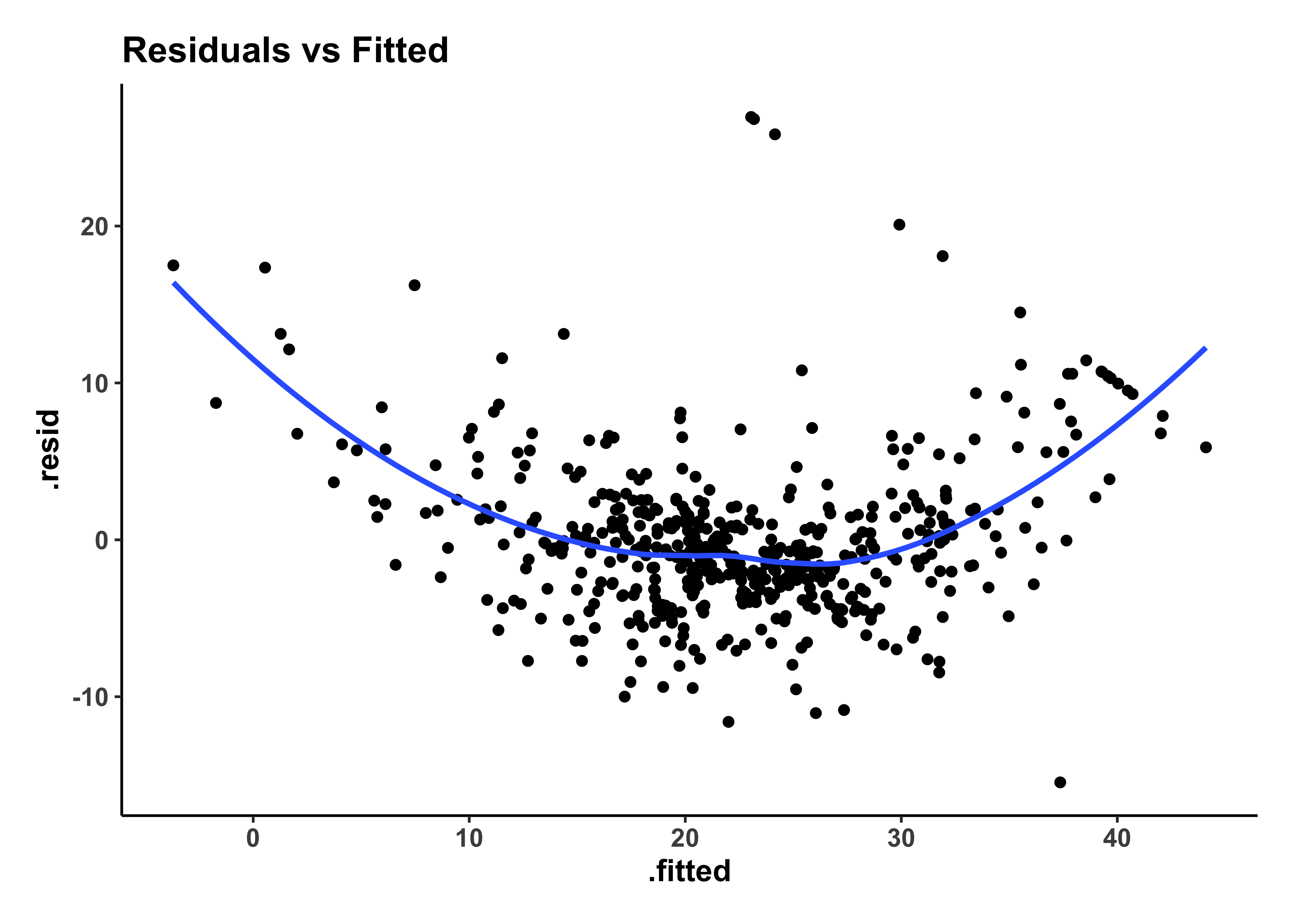
housing_model_final_augment %>%
gf_point(.resid ~ .fitted, title = "Residuals vs Fitted") %>%
gf_smooth() %>%
gf_theme(my_theme)
housing_model_final_augment %>%
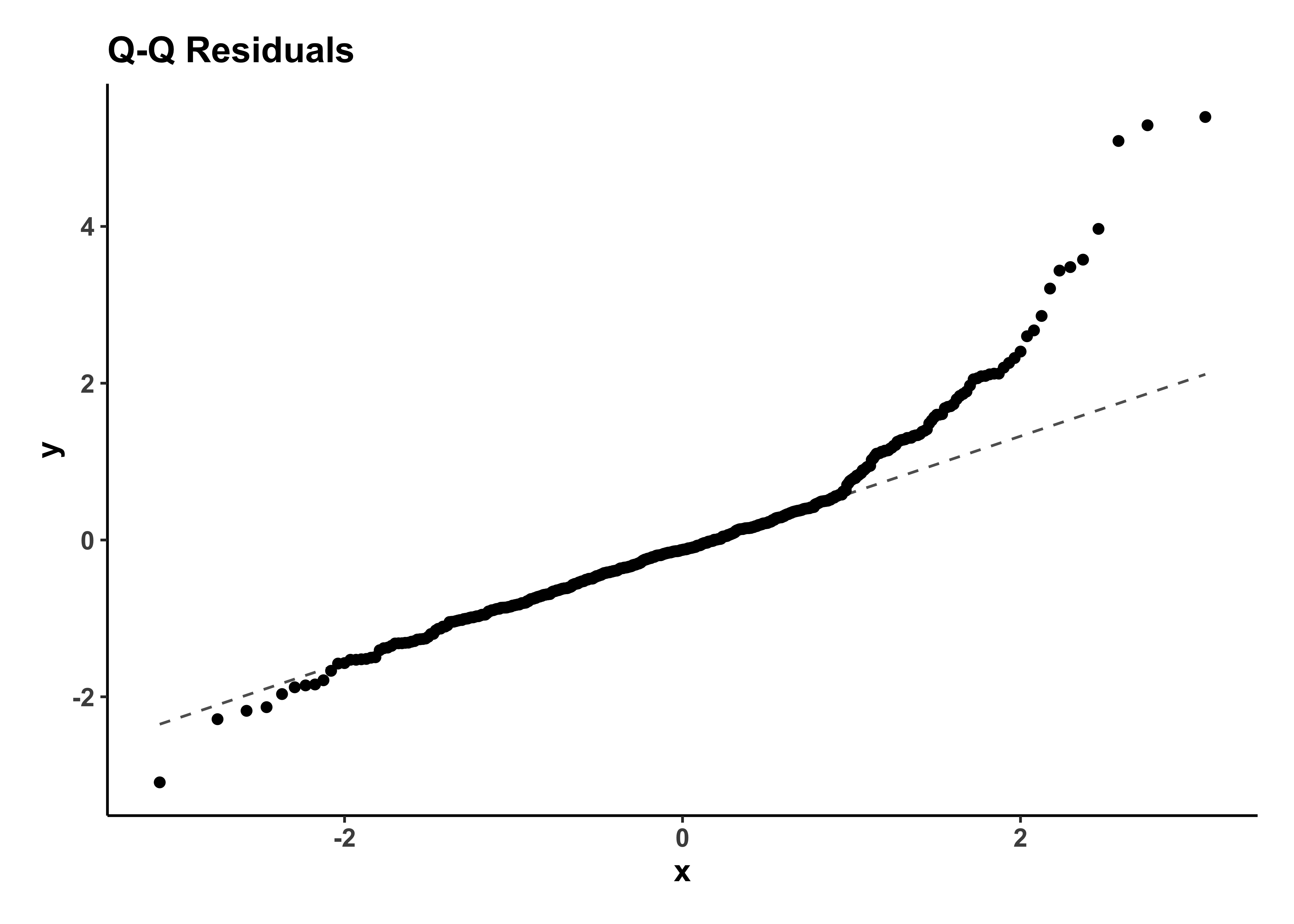
gf_qq(~.std.resid, title = "Q-Q Residuals") %>%
gf_qqline() %>%
gf_theme(my_theme)
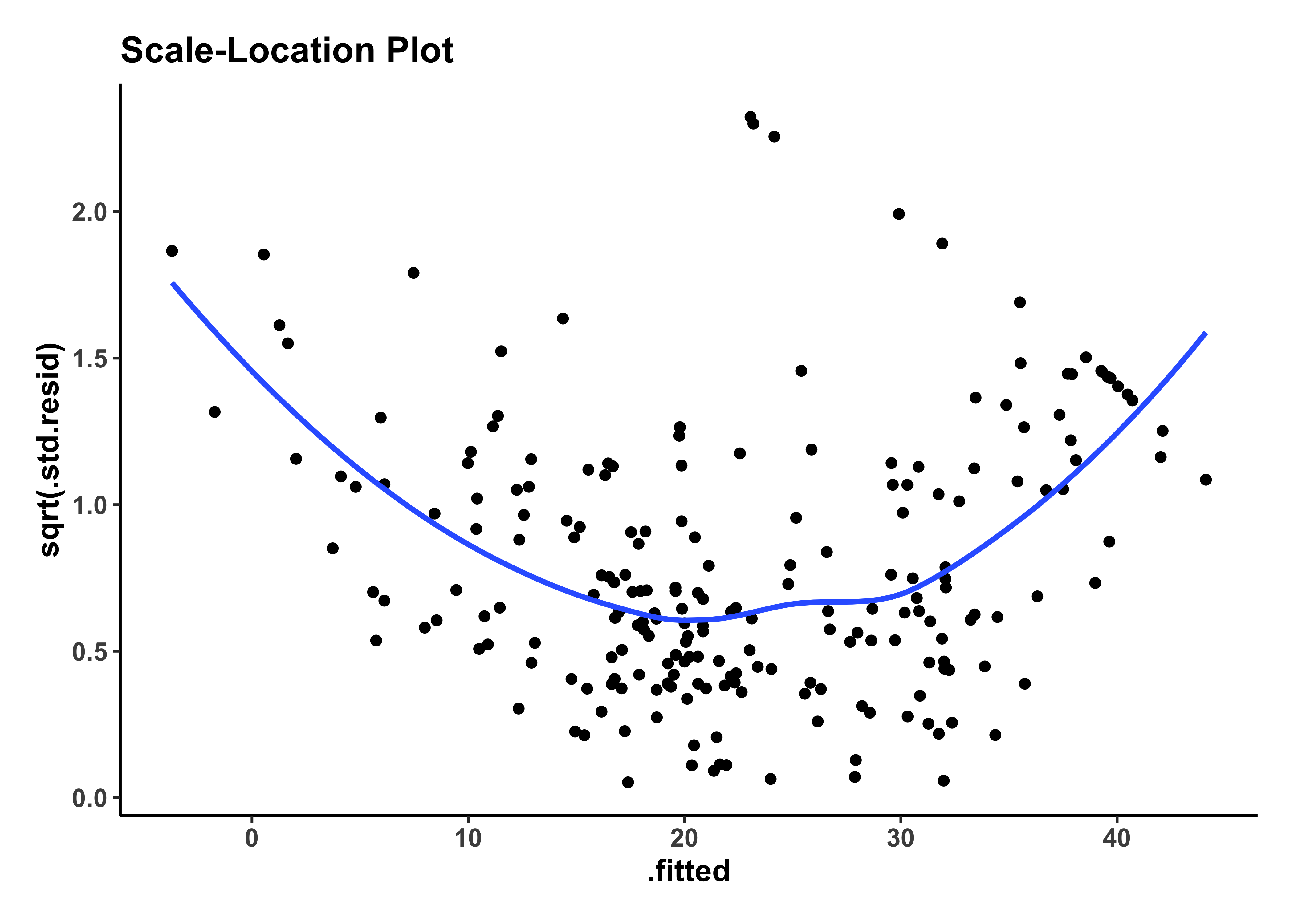
housing_model_final_augment %>%
gf_point(sqrt(.std.resid) ~ .fitted,
title = "Scale-Location Plot"
) %>%
gf_smooth() %>%
gf_theme(my_theme)
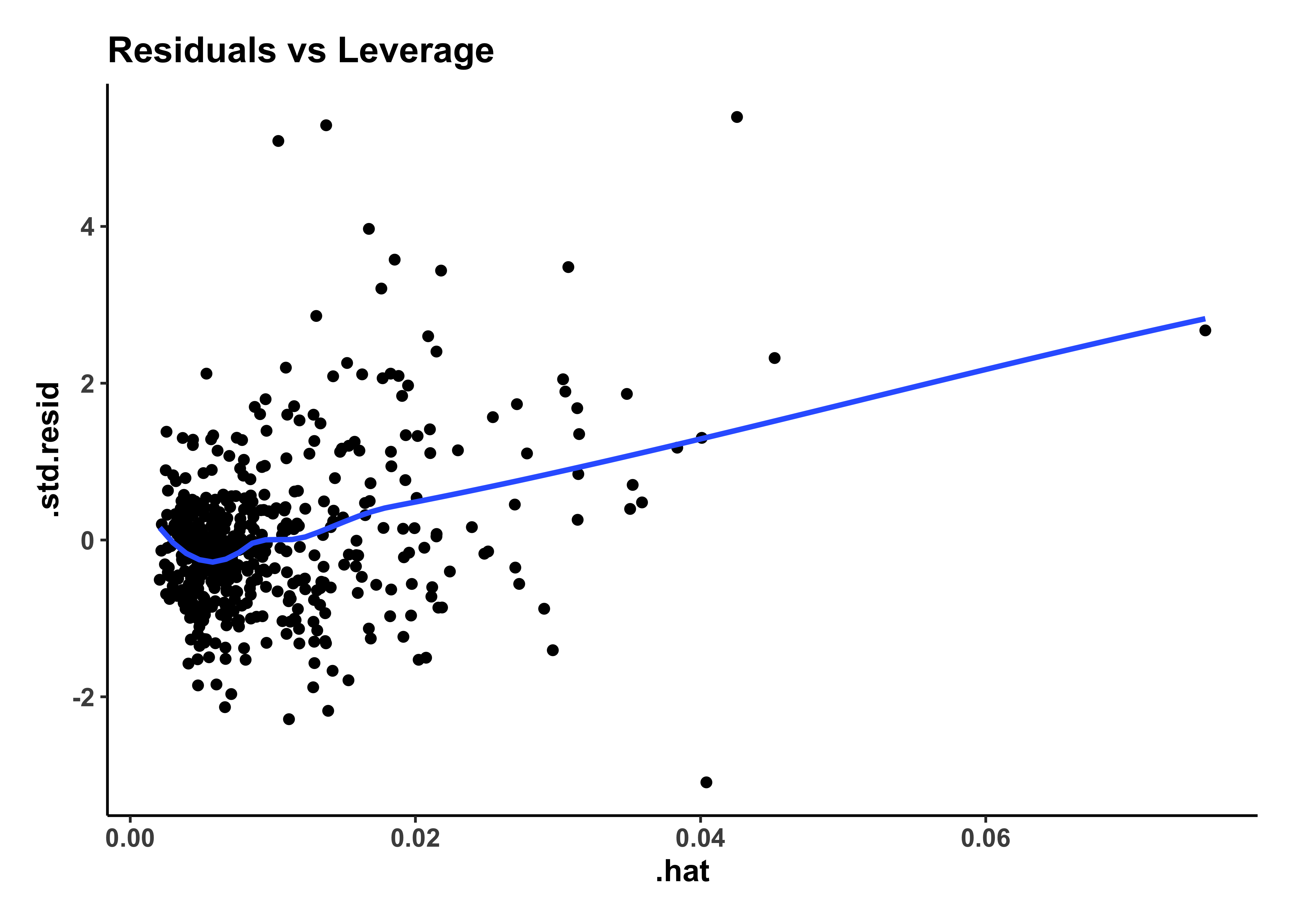
housing_model_final_augment %>%
gf_point(.std.resid ~ .hat,
title = "Residuals vs Leverage"
) %>%
gf_smooth() %>%
gf_theme(my_theme)The residuals plot shows a curved trend, and certainly does not resemble the stars at night, so it is possible that we have left out some possible richness in our model-making, a “systemic inadequacy”.
The Q-Q plot of residuals also shows a J-shape which indicates a non-normal distribution of residuals.
These could indicate that more complex model ( e.g. linear model with interactions between variables ( i.e. product terms ) may be necessary.
We have used a multiple-regression workflow that takes all predictor variables into account in a linear model, and then systematically simplified that model such that the performance was just adequate.
The models we chose were all linear of course, but without interaction terms : each predictor was used only for its main effect. When the diagnostic plots were examined, we did see some shortcomings in the model. This could be overcome with a more complex model. These might include selected interactions, transformations of target(
James, Witten, Hastie, Tibshirani, An Introduction to Statistical Learning. Chapter 3. Linear Regression. https://hastie.su.domains/ISLR2/Labs/Rmarkdown_Notebooks/Ch3-linreg-lab.html
Footnotes
James, Witten, Hastie, Tibshirani,An Introduction to Statistical Learning. Chapter 3. Linear Regression https://hastie.su.domains/ISLR2/Labs/Rmarkdown_Notebooks/Ch3-linreg-lab.html↩︎
Citation
@online{v.2023,
author = {V., Arvind},
title = {Tutorial: {Multiple} {Linear} {Regression} with {Backward}
{Selection}},
date = {2023-05-03},
url = {https://av-quarto.netlify.app/content/courses/Analytics/Modelling/Modules/LinReg/files/backward-selection-1.html},
langid = {en},
abstract = {Using Multiple Regression to predict Quantitative Target
Variables}
}